
How to use GCP and AWS big data and AI cloud services from Jupyter Notebook
How does a data scientist typically perform their standard job stack such as data processing and training models? In the case of playing around with test data or models, all the work can be done locally. But when that isn’t the case, data specialists often resort to the help of cloud solutions, which requires a lot of additional knowledge about SDK cloud libraries, command-line tools, and code deployment.
Typically a user can perform data processing or training jobs on cloud clusters using CLI tools either from the command line or in scripts and other automations. Another option is using a SDK for the language of choice. It enables developers to create and manage cloud services and provides an API as well as access to services in scripts and other automations.
It is also quite often necessary to get information about both running and completed jobs. Users can do it using the same tools or via the cloud console. Using different tools for completing one task can be quite inconvenient so having an instrument that allows data specialists to perform multiple tasks and access desired data and results without having to leave a familiar work environment can be very practical and time-saving.
What is the main tool of a data specialist? As a rule it is Jupyter Notebook. Jupyter Notebook is an open-source web application that allows you to create and share documents that contain live code, equations, visualizations, and narrative text. Uses include data cleaning and transformation, numerical simulation, statistical modeling, data visualization, machine learning, and much more.
A data science engineer’s role includes performing exploratory data analysis, data preparation, and training processes. The IPython kernel is the Python execution backend for Jupyter. Magics are instruments that are specific to and provided by the IPython kernel. There are various available Magics functions that IPython ships with. You can create and register your own Magics with IPython.
ML-DSL is an open source machine learning library developed to simplify the data specialist’s experience of interacting with cloud platforms such as Amazon AWS and the Google Cloud Platform. It lets data scientists and data analysts configure and execute ML/DS pipelines.
The main idea of ML-DSL is performing the standard data specialist’s job flow on cloud platforms from a Jupyter Notebook. Using specific Magics, you can prepare scripts to run spark jobs, train and deploy models, and get deployed model’s predictions.
The following features are currently available:
- Configuring and executing spark jobs for data processing on Google Dataproc and Amazon EMR.
- Configuring and executing ML/DS pipelines for training and deployment of models on Google AI Platform and Amazon SageMaker using the ML-DSL API.
- Configuring and executing ML/DS pipelines for data processing and training and deployment of models using Jupyter Notebook Magic functions.
ML-DSL overview
The ML-DSL provides a unified API and is responsible for interacting with cloud resources such as storages, distributed data processing, and calculations services that use Apache Hadoop and Spark stacks. It also provides machine learning platforms to build, train, and deploy machine learning models.
There are two implementations:
- Amazon
- Storage - Amazon S3
- Data processing - Amazon EMR
- Machine learning - Amazon Sagemaker
- Storage - Google Cloud Storage
- Data processing - Google Dataproc
- Machine learning - Google AI Platform
There is a set of profile classes that let you define all the necessary information for submitting and running jobs on cloud clusters and platforms: name of cluster, region, default bucket on cloud storage, and job-specific features such as job name and arguments.
Another important part of the ML-DSL API is the class of Jupyter “magic” functions. They allow specialists to define what an action will be doing:
- %py_script - register it’s cell content as a task script and save it as a python file with specified path and filename
- %py_script_open - register content of the specified python script as a task script.
- %py_load - loads content of python script to the cell
- %py_data - start a data processing job with specified parameters
- %py_train - submit and execute a training job with specified parameters
- %py_deploy - deploy your trained model on the ML platform
- %py_test - get online predictions from model resources for test data
- %logging - get logs of the specified job and print them out (currently available for Dataproc jobs)
There is also a set of shortcuts that is useful to define the details of a job. For example -e (--exec) for executing script immediately as a Jupyter cell (for testing purposes) or -n (--name) defines the name of a python script.
In this article, we are going to walk you through the experience of using ML DSL to execute an end-to-end data science process of data preparation, model training, and model serving in the cloud directly from a Jupyter Notebook.
ML-DSL for data scientists
Imagine you are a data scientist and you have a dataset. It is a corpus of some reviews (movie reviews for instance) and you want to build a sentiment analysis model (LSTM neural network) to classify reviews as positive or negative. The dataset is large and is located on cloud storage (Google Storage or Amazon S3), but you also have a small part of it stored locally.
First, you need to of course clean and process the data for further usage. You have written several functions in Jupyter Notebook for reading, cleaning, and using some pretrained word embeddings.
Import modules:
from pyspark import SQLContext, SparkContext
from pyspark.sql.window import Window
from pyspark.sql import Row
from pyspark.sql.types import StringType, ArrayType, IntegerType, FloatType
from pyspark.ml.feature import Tokenizer
import pyspark.sql.functions as F
Read glove.6B.50d.txt using pyspark:
def read_glove_vecs(glove_file, output_path):
rdd = sc.textFile(glove_file)
row = Row("glovevector")
df = rdd.map(row).toDF()
split_col = F.split(F.col('glovevector'), " ")
df = df.withColumn('word', split_col.getItem(0))
df = df.withColumn('splitted', split_col)
vec_udf = F.udf(lambda row: [float(i) for i in row[1:]], ArrayType(FloatType()))
df = df.withColumn('vec', vec_udf(F.col('splitted')))
df = df.drop('splitted', "glovevector")
w = Window.orderBy(["word"])
qdf = df.withColumn('vec', F.concat_ws(',', 'vec')).withColumn("id", F.row_number().over(w))
path = '{}/words'.format(output_path)
qdf.coalesce(1).write.format('csv').option("sep", "\t").option('header', 'true').save(path)
list_words = list(map(lambda row: row.asDict(), qdf.collect()))
word_to_vec_map = {item['word']: item['vec'] for item in list_words}
words_to_index = {item['word']: item["id"] for item in list_words}
return words_to_index, word_to_vec_map
Function to read review, tokenize each one, and replace words with indices in GloVe:
def prepare_df(path, const, words_dct):
rdd = sc.textFile(path)
row = Row("review")
df = rdd.map(row).toDF()
# Tokenize text
tokenizer = Tokenizer(inputCol='review', outputCol='words_token')
df_words = tokenizer.transform(df).select('words_token')
# Replace word with it's index
word_udf = F.udf(
lambda row: [words_to_index[w] if w in words_to_index.keys()
else words_to_index["unk"] for w in row],
ArrayType(IntegerType()))
df_stemmed = df_words.withColumn('words', word_udf(F.col('words_token')))
return df_stemmed.withColumn("class", F.lit(const))
Join together positive and negative reviews in the train/test dataset and save it for further work:
def save_dataset(df_pos, df_neg, path):
df = df_pos.union(df_neg)
w = Window.orderBy(["words"])
df = df.withColumn("review_id", F.row_number().over(w)).withColumn('int_seq',
F.concat_ws(',', 'words_stemmed'))
qdf = df.select(['review_id', 'int_seq', 'class'])
qdf.coalesce(1).write.format('csv').option('header', 'true').save(path)
And finally you can use a local sample of your dataset to prepare, train/test the dataset and save it:
sc = SparkContext(appName="word_tokenizer").getOrCreate()
sql = SQLContext(sc)
words_to_index, index_to_words, word_to_vec_map = read_glove_vecs("demo/data/glove.6B.50d.txt",
"output/words")
df_pos = prepare_df(f"{train_data}/pos/*.txt", 1, words_to_index)
df_neg = prepare_df(f"{train_data}/neg/*.txt", 0, words_to_index)
train_path = "output/train"
save_dataset(df_pos, df_neg, train_path)
df_pos = prepare_df(f"{test_data}/pos/*.txt", 1, words_to_index)
df_neg = prepare_df(f"{test_data}/neg/*.txt", 0, words_to_index)
test_path = "output/test"
save_dataset(df_pos, df_neg, test_path)
It works as you expected and you have prepared the dataset. You also prepare code in the Jupyter Notebook for training your LSTM model on this data.
Import the python libraries you need here:
import pandas as pd
import numpy as np
from tensorflow.python.lib.io import file_io
from tensorflow.keras.layers import Dense, Input, LSTM, Embedding, Dropout, Activation
from tensorflow.keras.models import Model
from tensorflow.keras.callbacks import Callback
from tensorflow.keras.optimizers import Adam
import matplotlib.pyplot as plt
import seaborn as sns
You have to load the training dataset and word embeddings:
def read_glove_vectors(glove_file):
files = file_io.get_matching_files('{}/part*'.format(glove_file))
for file in files:
with file_io.FileIO(file, 'r') as f:
word_to_vec_map = {}
words_to_index = {}
fl = f.readline()
for line in f:
line = line.strip().split('\t')
word_to_vec_map[line[0]] = np.array(line[1].split(','), dtype=np.float64)
words_to_index[line[0]] = int(line[2])
return words_to_index, word_to_vec_map
def read_csv(path):
files = file_io.get_matching_files('{}/part*'.format(path))
pdf = []
for file in files:
with file_io.FileIO(file, 'r') as f:
df = pd.read_csv(f)
if df is not None and len(df) != 0:
pdf.append(df)
if len(pdf) == 0:
return None
return pd.concat(pdf, axis=0, ignore_index=True).reset_index()
Functions to build your neural network:
def pretrained_embed_layer(word_to_vec_map, word_to_index, emb_dim):
emb_matrix = np.zeros((len(word_to_index) + 1, emb_dim))
for word, idx in word_to_index.items():
emb_matrix[idx, :] = word_to_vec_map[word]
return emb_matrix
def define_model(input_shape, emb_matrix, vocab_len, emb_dim, rnn_units, dropout=0.5):
sentence_indices = Input(input_shape, dtype="int32")
# Create the embedding layer pretrained with GloVe Vectors
embedding_layer = Embedding(input_dim=vocab_len, trainable=False, output_dim=emb_dim)
embedding_layer.build((None,))
embedding_layer.set_weights([emb_matrix])
# Propagate sentence_indices through your embedding layer
embeddings = embedding_layer(sentence_indices)
X = LSTM(units=rnn_units, return_sequences=False)(embeddings)
# Add dropout with a probability
X = Dropout(dropout)(X)
# Propagate X through a Dense layer
X = Dense(2)(X)
# Add a softmax activation
X = Activation("softmax")(X)
# Create Model instance which converts sentence_indices into X.
model = Model(inputs=sentence_indices, outputs=X)
return model
You also want to track losses and metrics and save charts with these:
class MetricCallback(Callback):
def on_train_begin(self, logs={}):
self.losses = []
self.accuracies = []
def on_batch_end(self, batch, logs={}):
self.losses.append(logs.get('loss'))
self.accuracies.append(logs.get('acc'))
def plot_metrics(callback, dir_to_save):
f, axes = plt.subplots(1, 2, figsize=(18, 5))
sns.lineplot(x=range(len(callback.losses)), y=callback.losses, ax=axes[0])
axes[0].title.set_text("Loss")
sns.lineplot(x=range(len(callback.accuracies)), y=callback.accuracies, ax=axes[1])
axes[1].title.set_text("Accuracy")
plt.tight_layout(.5)
plt.savefig('{}'.format(dir_to_save))
plt.show()
And finally train the model:
word_to_index, word_to_vec_map = read_glove_vectors("output/words/")
train_x, train_y, l = prepare_dataset("output/train/", word_to_index)
NUM_EPOCS = 3
RNN_STATE_DIM = 32
LEARNING_RATE = 0.01
vocab_len = len(word_to_index) + 1
emb_dim = word_to_vec_map["cucumber"].shape[0]
emb_matrix = pretrained_embed_layer(word_to_vec_map, word_to_index, emb_dim)
model = define_model((N,), emb_matrix, vocab_len, emb_dim, RNN_STATE_DIM)
print(model.summary())
model.compile(loss='binary_crossentropy', optimizer=Adam(lr=LEARNING_RATE),
metrics=['accuracy'])
# fit model
metrics = MetricCallback()
a = model.fit(train_x, train_y, batch_size=1024, epochs=NUM_EPOCS,
callbacks=[metrics], shuffle=True)
# save the model to file
model_dir = "output/movie_model"
file_io.recursive_create_dir(model_dir)
path = f'{model_dir}/saved_model'
tf.saved_model.save(model, path)
path_chart = '{}/metrics.png'.format(model_dir)
plot_metrics(metrics, local_path_chart)
It works well, returns training progress, and saves the model to a specified path. You’ve just prepared a dataset and trained the model locally on a small part of your dataset.
But the full dataset is huge and you would like to use it for training your model. You need a more powerful instance or multiple instances than your laptop to process the whole dataset and train the model.
The best idea is delegating a data processing job to a cloud cluster that uses Apache Spark or another open-source big data framework to process and analyze the data. This is a Dataproc for Google Cloud Platform or Amazon EMR for Amazon Web Services.

But how to organize it? You know you have credentials to interact with Google Cloud Platform services and there is a Dataproc cluster you can use for your task. So in fact you have all the information you need:
- Cluster name “cluster”,
- Name of project “project”,
- Region “central”,
- Google Cloud Storage bucket “bucket”, where your dataset is saved
- JSON with credentials (you can set them as an environment variable GOOGLE_APPLICATION_CREDENTIALS).
So how do you go about submitting the job to Dataproc? As a general rule this can be done by using the console, command-line tools, or client libraries. So your code has to be organized as a python script as well as with all additional code. Then by going to the Dataproc console or by starting a specific command in the shell or by writing additional python code you can submit the job to the cluster.
Data processing
Using ML-DSL Magics you can do all necessary procedures in Jupyter Notebook.
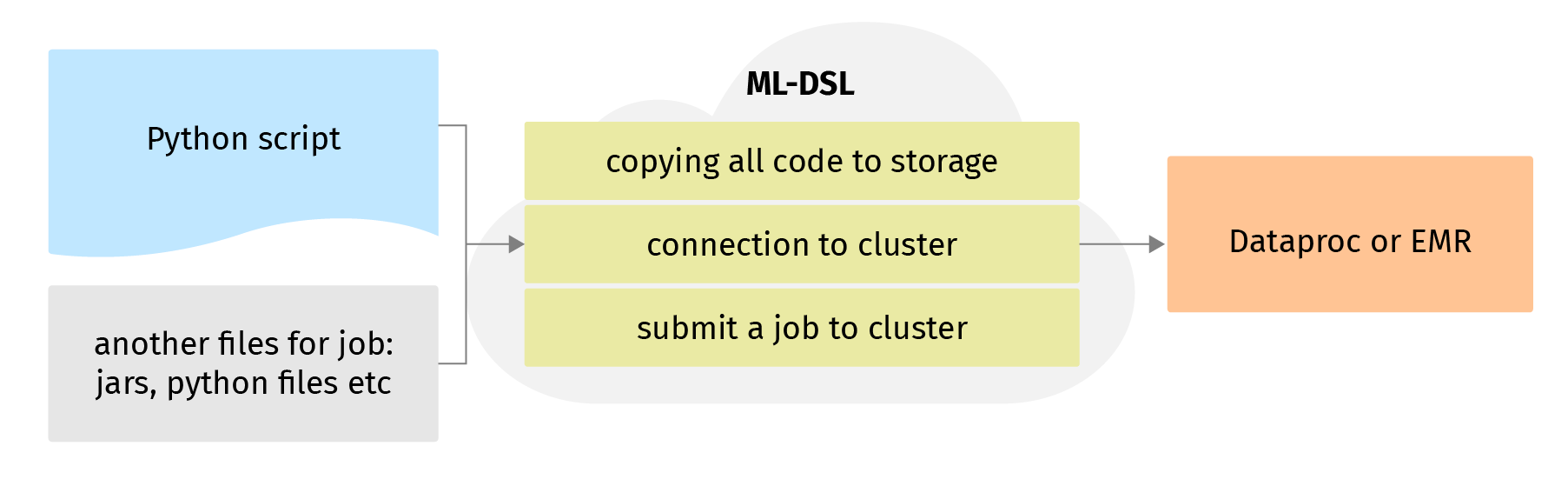
First, you can organize your code for a pyspark job by using one of the ML-DSL Magics: %py_script, %py_script_open or %py_load in your notebook. The commands save your code as a python file or load it from an already existing script. You can also run code locally with these Magics for testing.
%%py_script -e -n text_tokenizer.py -p demo/scripts --train_path aclImdb/train --test_path aclImdb/test --word_embeds aclImdb/glove.6B.50d.txt - o demo/output
# !/usr/bin/python
from pyspark import SQLContext, SparkContext
from pyspark.sql.window import Window
import pyspark.sql.functions as F
from pyspark.sql import Row
from pyspark.sql.types import StringType, ArrayType, IntegerType, FloatType
from pyspark.ml.feature import Tokenizer
def prepare_df(path, const, words_dct):
rdd = sc.textFile(path)
row = Row("review")
df = rdd.map(row).toDF()
# Tokenize text
tokenizer = Tokenizer(inputCol='review', outputCol='words_token')
df_words = tokenizer.transform(df).select('words_token')
# Replace word with it's index
word_udf = F.udf(
lambda row: [words_to_index[w] if w in words_to_index.keys()
else words_to_index["unk"] for w in row],
ArrayType(IntegerType()))
df_stemmed = df_words.withColumn('words', word_udf(F.col('words_token')))
return df_stemmed.withColumn("class", F.lit(const))
...
if __name__ == '__main__':
parser = argparse.ArgumentParser()
parser.add_argument('--train_path', type=str, help="train positive reviews path")
parser.add_argument('--test_path', type=str, help="test positive reviews path")
parser.add_argument('--word_embeds', type=str, help="Path to glove word embeddings")
parser.add_argument('--output_path', type=str, help="Sequences output path")
args, d = parser.parse_known_args()
output_path = args.output_path
sc = SparkContext(appName="word_tokenizer").getOrCreate()
sql = SQLContext(sc)
words_to_index, index_to_words, word_to_vec_map = read_glove_vecs(args.word_embeds,
output_path)
df_pos = prepare_df(f"{args.train_path}/pos/*.txt", 1, words_to_index)
df_neg = prepare_df(f"{args.train_path}/neg/*.txt", 0, words_to_index)
train_path = '{}/train'.format(output_path)
save_dataset(df_pos, df_neg, train_path)
print('Train saved to: "{}"'.format(train_path))
df_pos = prepare_df(f"{args.test_path}/pos/*.txt", 1, words_to_index)
df_neg = prepare_df(f"{args.test_path}/neg/*.txt", 0, words_to_index)
test_path = '{}/test'.format(output_path)
save_dataset(df_pos, df_neg, test_path)
print('Test saved to: "{}"'.format(test_path))

The next step is providing all the information and credentials you need to your task. It is necessary in order to be able to submit the job. With ML-DSL it is possible to reach that by instantiating a PySparkJobProfile class. The class has attributes that describe the information you need to interact with the Cloud Platform: cluster, project (in case of GCP), region, and bucket name.
You also define specific attributes of the class: arguments, if there are any for your script, lists of files, jars, archives, and packages when it is necessary. You also have to define the name prefix of your job, for example “word_tokenizer” or “review_processing”. It is beneficial to use meaningful job names.
The last step is to specify the cloud platform (Google Cloud Platform in our case). There is a specific ML-DSL class Platform for this.
platform = Platform.GCP
profile = PySparkJobProfile(root_path='demo/scripts',
bucket='your_bucket',
project='your_project',
cluster='your_cluster',
region='global',
ai_region='us-central1-a',
job_prefix='demo_job',
job_async=False)
profile.args = profile.load_profile_data("demo/spark_job_args_gcp.json")
PySparkJobProfile.set('JobProfile', profile)
And that’s it. You can finally start the job using ML-DSL Magic %py_data. Just add your PySparkJobProfile instance, platform instance, name of your python script, and output path (where the processed data will be saved) as arguments to this Magic and run the cell. ML-DSL will handle all the job tasks: prepare an instance of the spark job with your arguments, make a connection session to the cloud platform, and submit the job to the Dataproc cluster you’ve defined as the profile instance attribute cluster:
%py_data -n text_tokenizer.py -p JobProfile -pm $platform -o gs://your_bucket/mldsl/data

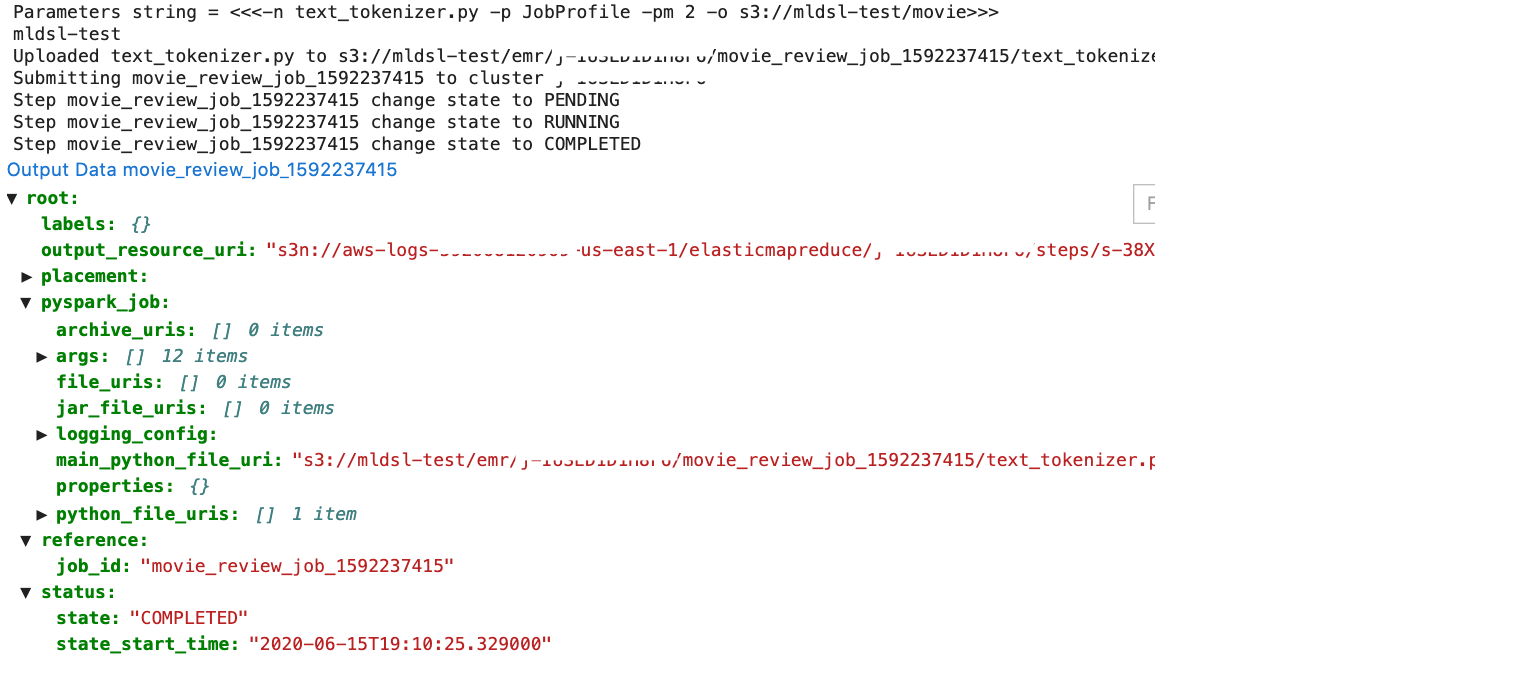
During execution, ML-DSL returns general information about the job description and status as well as links for the output path and to the logs of the job on the cloud platform to the output of the running cell.
However, what if you have access to the Amazon EMR cluster and want to run your job there? There are not a lot of changes required. You only need to redefine the PySparkJobProfile object and platform with the parameters specific to the case.
platform = Platform.AWS
profile = PySparkJobProfile(bucket='mldsl-bucket',
cluster='mldsl-cluster',
region='global',
job_prefix='movie_review_job',
root_path='demo/scripts', project=None, ai_region='us-east-1',
job_async=False)
profile.args = profile.load_profile_data("demo/spark_job_args_aws.json")
profile.packages = ["com.amazonaws:aws-java-sdk-pom:1.10.34", "org.apache.hadoop:hadoop-aws:2.6.0"]
PySparkJobProfile.set('JobProfile', profile)
The command and it’s arguments for submitting the job to an EMR cluster are the same as to Dataproc. Actually ML-DSL does the same process: prepare an instance of the spark job with your arguments, make a connection session to the cloud platform, and submit the job. As you would expect, the final result is fairly similar (except that the data and output path is not on Google Cloud Storage but on S3).
%py_data -n text_tokenizer.py -p JobProfile -pm $platform -o s3: //mldsl-test/movie
Training the model
So you’ve now successfully prepared your data on the cloud service without having to leave your working notebook. Now It’s time to train your sentiment classifier. You decided to train a simple LSTM model and, of course, want to train your model on a full dataset. The best option here is to run the training on the Google AI Platform.
The standard process of running a training job on the platform includes such steps as: creating a python file with the model, adding code for downloading data from Google Cloud Storage so ML Platform can use it, adding code to export and save the model after the training is finished, and preparing a training application package and submitting the training job to the platform. This last step can be done by using the console, command-line tools, or client libraries).
You prepare your code so it contains everything described above: importing required libraries, downloading data, and training and exporting the model. So just like with data processing, you can organize this code to script, save it and test on local data using one of the ML-DSL Magics: %py_script, %py_script_open or %py_load.
%%py_script -n mr_model.py -p demo/model/words/trainer -o dev/models --epochs 3 --train_path gs://your_bucket/data/train --word_embeds gs://your_bucket/data/words
# !/usr/bin/python
import pandas as pd
import numpy as np
from tensorflow.python.lib.io import file_io
from tensorflow.keras.layers import Dense, Input, LSTM, Embedding, Dropout, Activation
from tensorflow.keras.models import Model
from tensorflow.keras.callbacks import Callback
...
def define_model(input_shape, emb_matrix, vocab_len, emb_dim, rnn_units, dropout=0.5):
sentence_indices = Input(input_shape, dtype="int32")
# Create the embedding layer pretrained with GloVe Vectors
embedding_layer = Embedding(input_dim=vocab_len, trainable=False, output_dim=emb_dim)
...
if __name__ == '__main__':
parser = argparse.ArgumentParser()
parser.add_argument('--train_path', type=str, help="Train files path")
parser.add_argument('--output_path', type=str, help="Models output path")
parser.add_argument('--word_embeds', type=str, help="Models output path")
parser.add_argument('--epochs', type=int, help="Number of epochs")
args, d = parser.parse_known_args()
...
model = define_model((N,), emb_matrix, vocab_len, emb_dim, RNN_STATE_DIM)
print(model.summary())
model.compile(loss='binary_crossentropy', optimizer=Adam(lr=LEARNING_RATE),
metrics=['accuracy'])
# fit model
metrics = MetricCallback()
a = model.fit(train_x, train_y, batch_size=1024, epochs=NUM_EPOCS,
callbacks=[metrics], shuffle=True)
...
tf.saved_model.save(model, remote_path)
The same way you define a setup.py file for packaging code for AI Platform:
%%py_script --name setup.py --path demo/model/words
# %py_load demo/model/words/setup.py
# !/usr/bin/python
from setuptools import setup, find_packages
REQUIRED_PACKAGES = ['Keras==2.0.4', 'matplotlib==2.2.4', 'seaborn==0.9.0']
setup(
name='trainer',
version='1.0',
packages=find_packages(),
install_requires=REQUIRED_PACKAGES,
author='Grid Dynamics ML Engineer',
author_email='griddynamics@griddynamics.com',
url='https://griddynamics.com'
)
To describe information required to submit the training job to the AI Platform, you should define the specific Profile and Platform GCP. In the case of AI PLatform instantiating, the AIProfile class helps. It has attributes as ai_region, scale_tier, runtime_version, python_version for the AI Platform Training service setting up resources for your job, package_name, and package_dst for packaging your code and your script’s arguments.
platform = Platform.GCP
profile = AIProfile(bucket='your_bucket',
cluster='your_cluster',
region='global',
job_prefix='train',
root_path='demo/model/words',
project='gd-gcp-techlead-experiments',
ai_region='us-central1',
job_async=False,
package_name='trainer',
package_dst='mldsl/packages',
runtime_version='1.15',
python_version='3.7')
profile.arguments = profile.load_profile_data("demo/train_args_gcp.json")
AIProfile.set('AIProfile', profile)
Now it’s time to train. You can start the task using ML-DSL Magic %py_train. You pass your AIProfile, platform, and name of your script as arguments for this Magic.
%py_train -n mr_model.py -p AIProfile -pm $platform -o gs://your_bucket/mldsl

ML-DSL takes all the steps related to packaging the code and submits it to AI Platform:
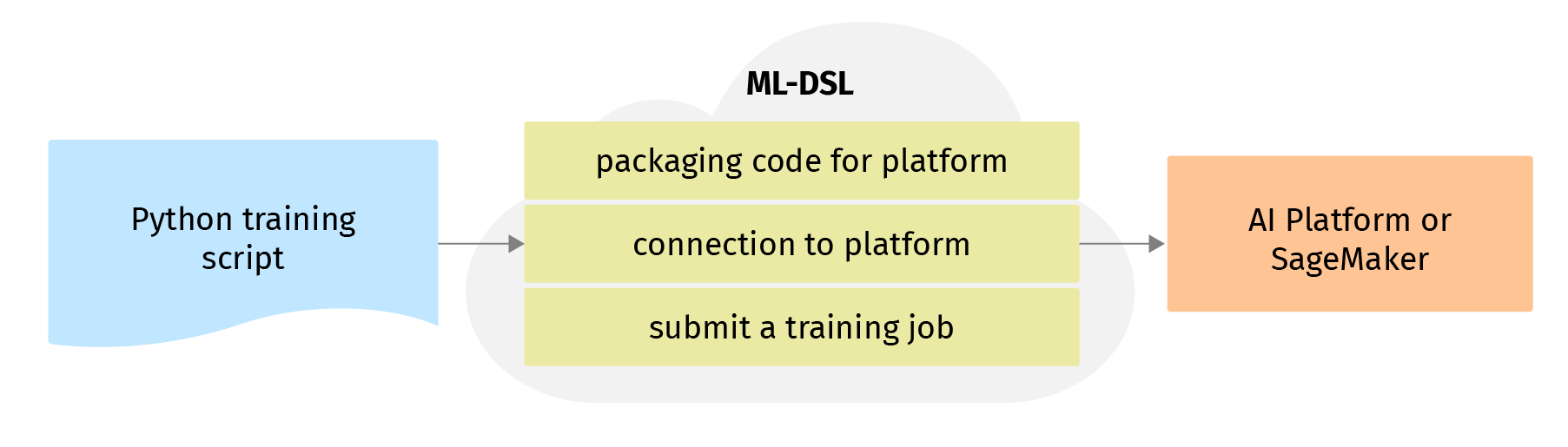
- Packages code and transfers it to a Cloud Storage bucket that your project can access and which you define in the AIProfile instance;
- Prepares a training job description for AIPlatform;
- Makes a connection session to the cloud platform and request services to set up resources for your job;
- Waits while the training service runs your application;
- Returns the status and information about the training process and results (place, where the trained model was saved, charts, price etc) after it finishes to the output of the running cell.

There are a couple of key differences if you want to run your training job on Amazon SageMaker. Some inputs for the training script need to be defined using specific environment variables such as SM_CHANNEL_TRAINING, SM_MODEL_DIR etc. Instead of an instance of AIProfile, you create an object of the SageMakerProfile class. There are attributes specific to submitting a training job to SageMaker: type of container (XGBoost, PyTorch, TensorFlow etc), framework_version, instance_type, and intance_count for setting up service resources to run the job.
from sagemaker.pytorch import PyTorch
platform = Platform.AWS
profile = SageMakerProfile(bucket='mldsl-bucket',
cluster='mldsl-cluster',
region='us-east-1',
job_prefix='mldsl_test',
container=PyTorch,
root_path='demo/scripts/',
framework_version='1.4.0',
instance_type='ml.m4.xlarge',
use_cloud_engine_credentials='SageMakerRoleMlDsl')
SageMakerProfile.set('SageMakerProfile', profile)
The training process is the same as in the case of Google AI Platform using %py_train. You pass your AIProfile, platform, and name of your script.
%py_train -n pytorch_model.py -p SageMakerProfile -pm $platform -o s3://mldsl-bucket/movie/output --training s3://mldsl-bucket/movie/data
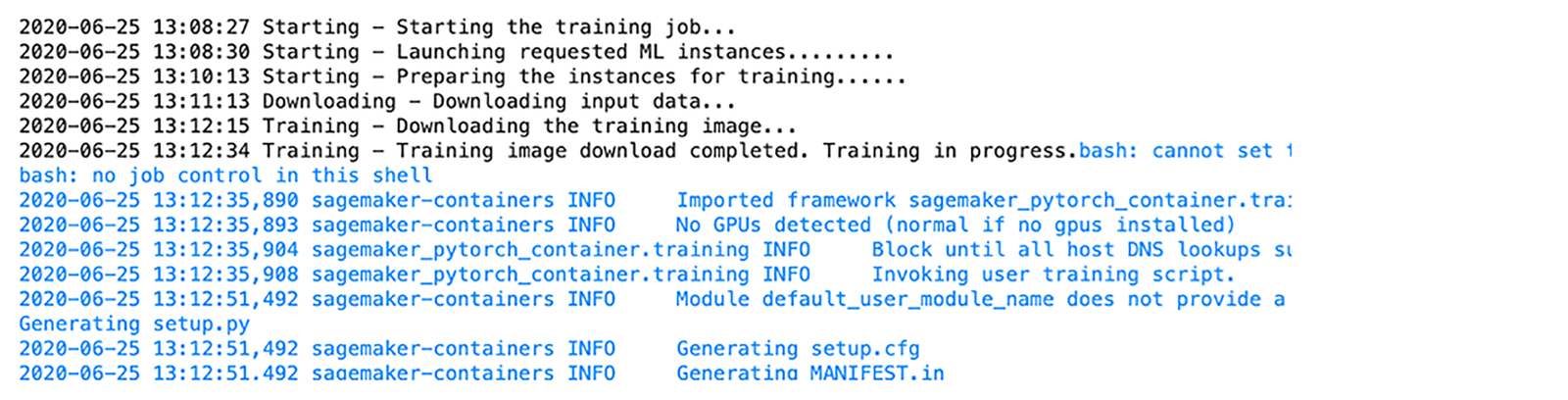
ML-DSL returns information about the training process and results after it finishes to the output of the running cell.
Deployment
The training job on SageMaker returns the result of a trained estimator that can be deployed to an endpoint where the model is available to provide inferences. ML-DSL provides it to the next Jupyter cell and you can run this cell and use it for deployment as shown on the picture below (using estimator method “deploy”).

# Use job_mldsl_test_1593109354 instance to browse job properties.
job_mldsl_test_1591265953 = job_tracker['mldsl_test_1591265953']
Deployment of model
predictor = job_mldsl_test_1591265953.deploy(initial_instance_count=1, instance_type='ml.m4.xlarge',
endpoint_name='mldsl-pytorch')
For deploying models on platforms you can create DeployAIProfile and SageMakerProfile for AI Platform and SageMaker respectively. It’s attributes let you include information about the saved model path, framework version, custom code, name of model endpoint, and version name.
platform = Platform.GCP
profile = DeployAIProfile(bucket='your_bucket',
cluster='your_cluster',
region='global',
job_prefix='deploy',
root_path='demo/deploy',
project='your_project',
ai_region='us-central1',
job_async=False,
runtime_version='1.15',
python_version='3.7',
version_name='v1',
is_new_model=True,
path_to_saved_model='gs://your_bucket/saved_model/')
profile.arguments = {
"framework": "TENSORFLOW"
}
DeployAIProfile.set('AIProfile', profile)
The deployment can be done using ML-DSL Magic %py_deploy. You send as arguments the name of the model, your profile instance, and platform.
%py_deploy -n mldsl_demo -p AIProfile -pm $platform

For deployment models on SageMaker, you can define the SageMakerProfile instance (just like for training) and run the cell with Magic %py-deploy.
% py_deploy -n pytorch_model.py -p TestDeploySMProfile -pm $platform

Here the arguments are the same with one key difference: you add the name of the python script that is the entry point (contains methods predict_fn, output_fn etc) and path where the script is located.
Getting inferences
The SageMaker deployment described above returns an estimator that can be used to provide inferences.
# Use job_mldsl_test_1593153514 instance to browse job properties.
predictor = job_tracker['mldsl_test_1591268290_predoctor']
PREDICTION
preds = predictor.predict(x_test)
prediction = preds.argmax(axis=1)
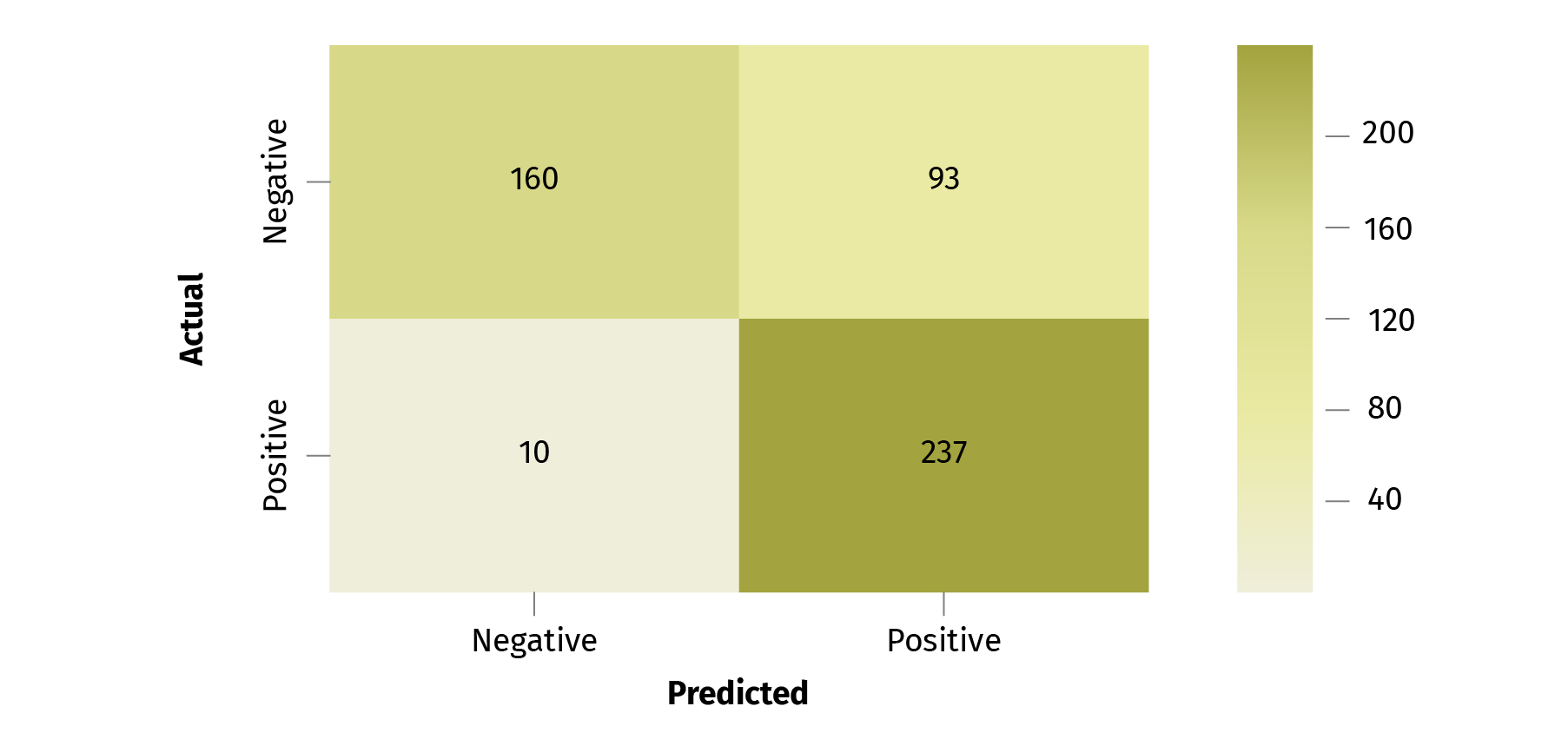
conf_mat = confusion_matrix(y, prediction)
fig, ax = plt.subplots(figsize=(8, 6))
classes = ['Negative', 'Positive']
sns.heatmap(conf_mat, annot=True, fmt='d', xticklabels=classes,
yticklabels=classes, cmap=plt.cm.BuGn)
plt.ylabel('Actual')
plt.xlabel('Predicted')
plt.show()
To get predictions of the model deployed on the AI Platform you can use the same DeployAIProfile profile as defined for the deployment process. Except that it’s necessary to prepare some test data and send it to ML-DSL %py_test magic as an argument as well as platform and profile instance:
%py_test -n mldsl -p AIProfile -pm $platform -t $test
activation = [[0.6321, 0.3678], [0.1271, 0.8728], [0.0241, 0.9758], [0.0035, 0.9964], [0.9954, 0.0045], ...,
[0.4544, 0.5455], [0.9880, 0.0119], [0.9955, 0.0044]]
preds = np.array(activation).argmax(axis=1)
target_names = ['negative review', 'positive review']
print(classification_report(test_y, press, target_names=target_names))

Running the cell returns output of the deployed model (in our case it is an output of a softmax layer in our neural network classifier) to the next cell. You can use this output to prepare some analytics about the model's quality: classification reports, confusion_matrix etc.
Congratulations! You have now completed a data scientist’s job circle, which includes running spark applications on cloud clusters (Dataproc and EMR) for data processing, training and deployment of a model using cloud services (AIPlatform and SageMaker), and getting inferences of the model in Jupyter Notebook. It’s good to have all the developments in one place so ML-DSL can be a very convenient tool for your job.
ML-DSL in Grid Dynamics’ projects
ML-DSL was used in Grid Dynamics’ Anomaly Detection project as part of its workflow design. It has two implementations:
- With Google Cloud Project: GCS, Dataproc, AI Platform
- With Amazon Cloud Services: S3, EMR, Sagemaker
There are two areas where ML-DSL performed its role:
- ML-DSL is the core library for the Jupyter Lab notebook, which consists of the data science pipeline for the anomaly detection machine learning models data preparation, design, training, evaluation, and publishing.
- ML-DSL is used as an entrypoint for the anomaly detection workflow deployment automation.
In the Amazon Cloud Services case, the CloudFormation was used for the Anomaly Detection solution deployment. Deployment manifests require Sagemaker endpoints and EMR Spark Streaming jobs deployment. The simple Lambda function was implemented, which consumed the ML-DSL library, simplifying and unifying all the related operations.
Conclusion
ML-DSL is a useful tool that can help data specialists to simplify the experience of interacting with cloud platforms and organize all working developments in Jupyter Notebooks. Currently it supports configuring and executing ML/DS pipelines on Google Cloud Platform and Amazon Web Services.
ML-DSL was used in several Grid Dynamics’ projects including the Unsupervised Real Time Anomaly Detection project.
ML-DSL will be published as open source. There are also many plans on how to further develop it including:
- Adding support for Microsoft Azure services;
- Developing new and expanding existing Profile classes;
- Adding support for creating custom docker containers in SageMaker to use your own algorithms and models.
References
- Unsupervised real time anomaly detection, Alex Rodin, white paper, Grid Dynamics Blog
- Grid Dynamics ML-DSL project, GitHub

